Data Science Applications
Bringing data to the end-users.
Deep Learning with Histologic Images
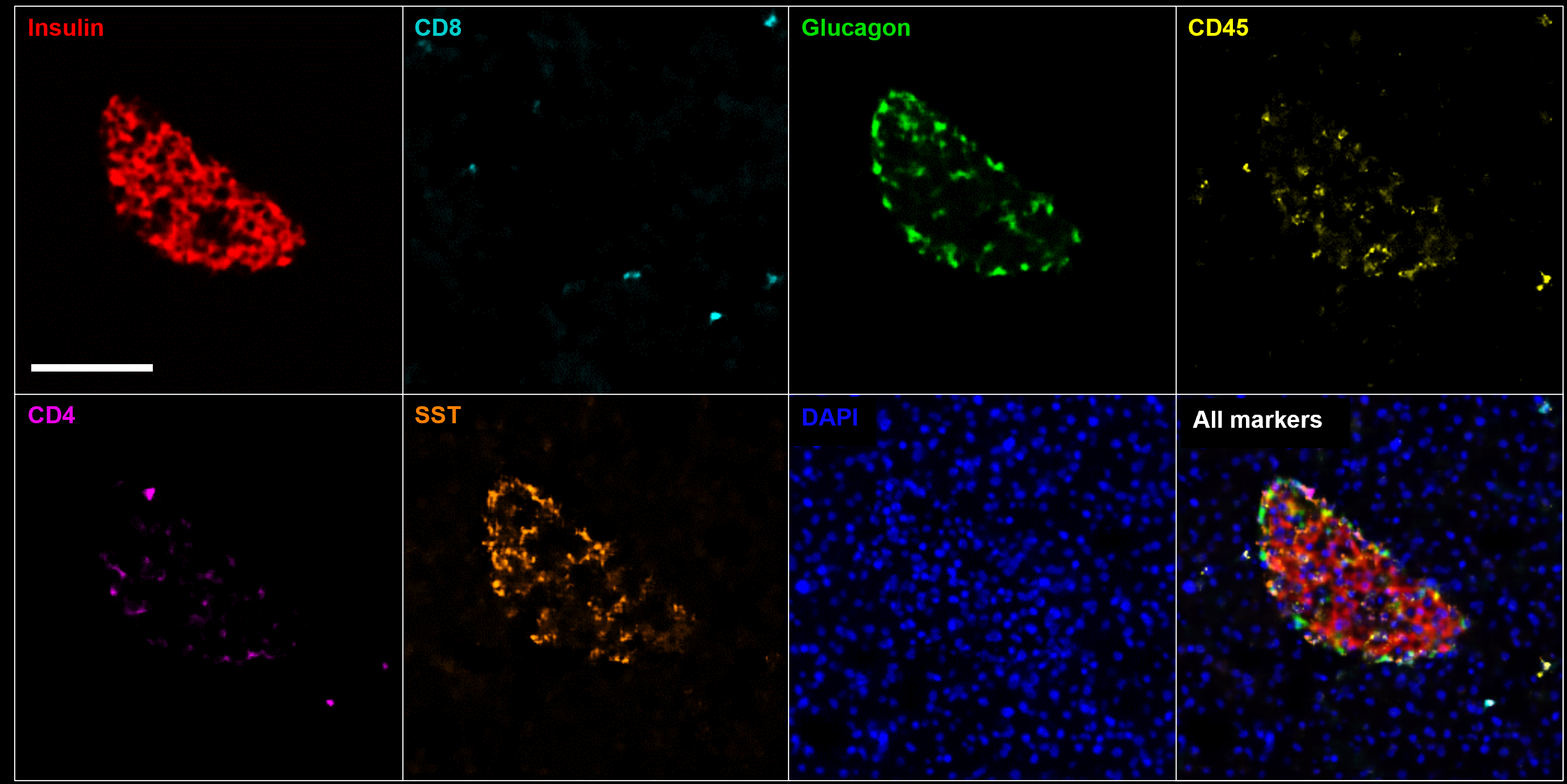
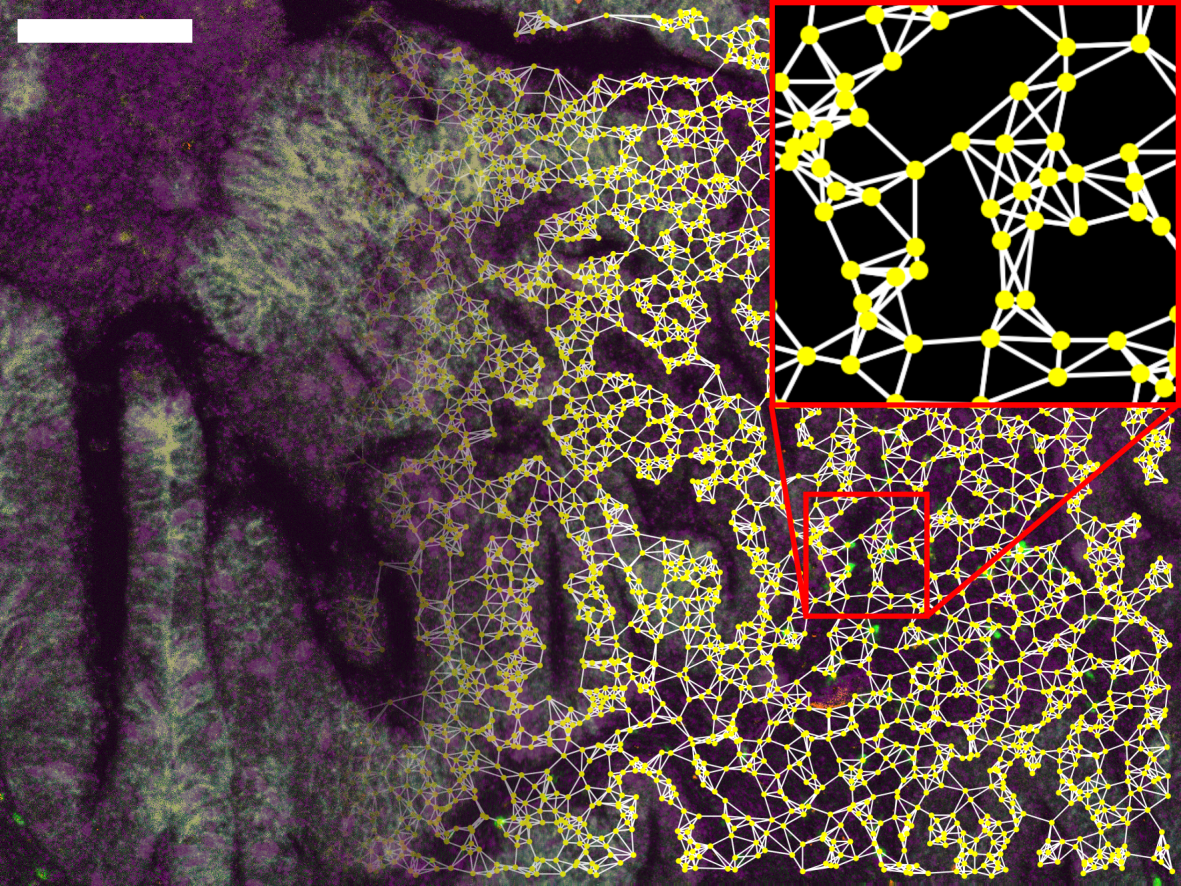
We use well-established convolutional neural network (CNN) architectures to create predictive image classification models on supervised training datasets. To this end, we can work with both weak (patient-level) annotations and strong (tissue or cell-level) annotations for the training data set. For whole-slide image data, we employ either image tiling or an in-house developed cell-based data representation (XXX link to Cell2Grid applicationXXX) to reduce the data size and burden on compute hardware.
We used this approach to predict the 5-year relapse risk of colon cancer patients from fm-IHC image crops stained with a marker panel targeting multiple immune cell phenotypes and tumor markers.
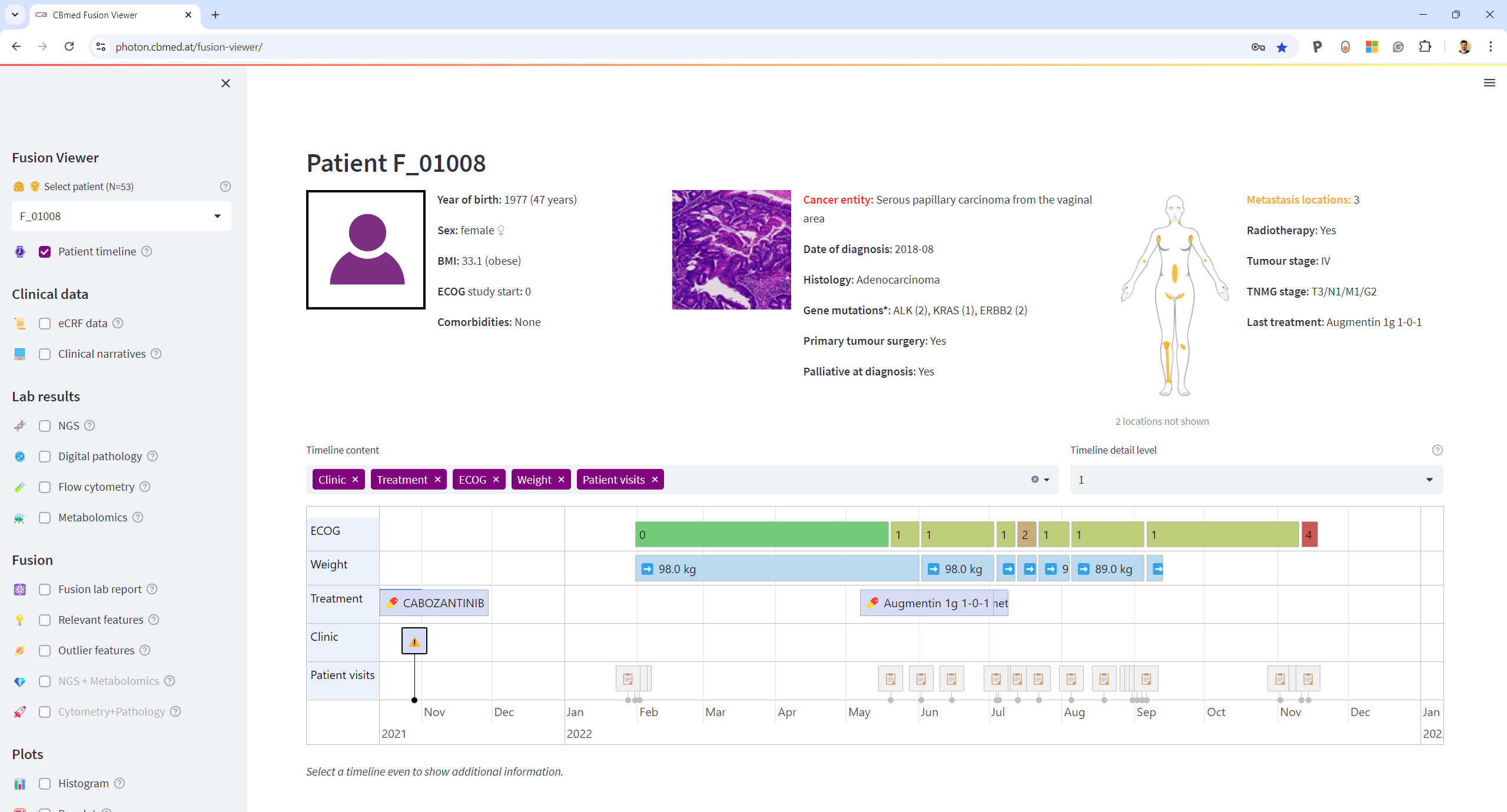
Molecular Tumor Board
Tumor boards are meetings of cancer experts, including oncologists, radiologists and other professions, to discuss the next treatment steps for cancer patients. To help them make informed decisions based on reliable data, our FusionViewer molecular tumor board aims to provide them with data and insights obtained from different biological layers of the patient, including NGS, metabolomics, flow cytometry, histologic imaging, and others. To complete the holistic picture of each patient, relevant clinical data features are extracted from unstructured electronic healthcare records (EHR) using natural language processing pipelines.
Compared to existing tumor board software solutions, FusionViewer integrates omics and non-omics data from multiple sources and shows them directly in conjunction with all available clinical data. This simplifies the analysis of cross-domain data both for oncologists during a tumor board and for researchers that aim and
We use on-premise hosted structured databases to store all data required for visualizations in a secure and anonymized way. Locally hosted web-applications access these databases to bring the data to the end-users through a simple, installation-free web interface.
Drug Screening Analysis Pipeline
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua enim sut.

Synthetic Data
Our goal is to become the leading expert center in the practical application of synthetic histologic images. This aims at making synthetic images useful for training knowledge extraction pipelines and to substitute real image data whenever a specialized hypothesis or research task demands the expansion of existing image data sets. Our methods range from procedural algorithms to machine-learning methods, depending on the size and availability of existing, real image data and the a priori knowledge about the image content. A first showcase of using a combination of procedural cell-based image content generation and a generative adversarial network for the generation of multiplex IHC images of murine pancreatic islets shows promising results of this approach for solving complex research questions.